Research and Projects
DeepCell Label
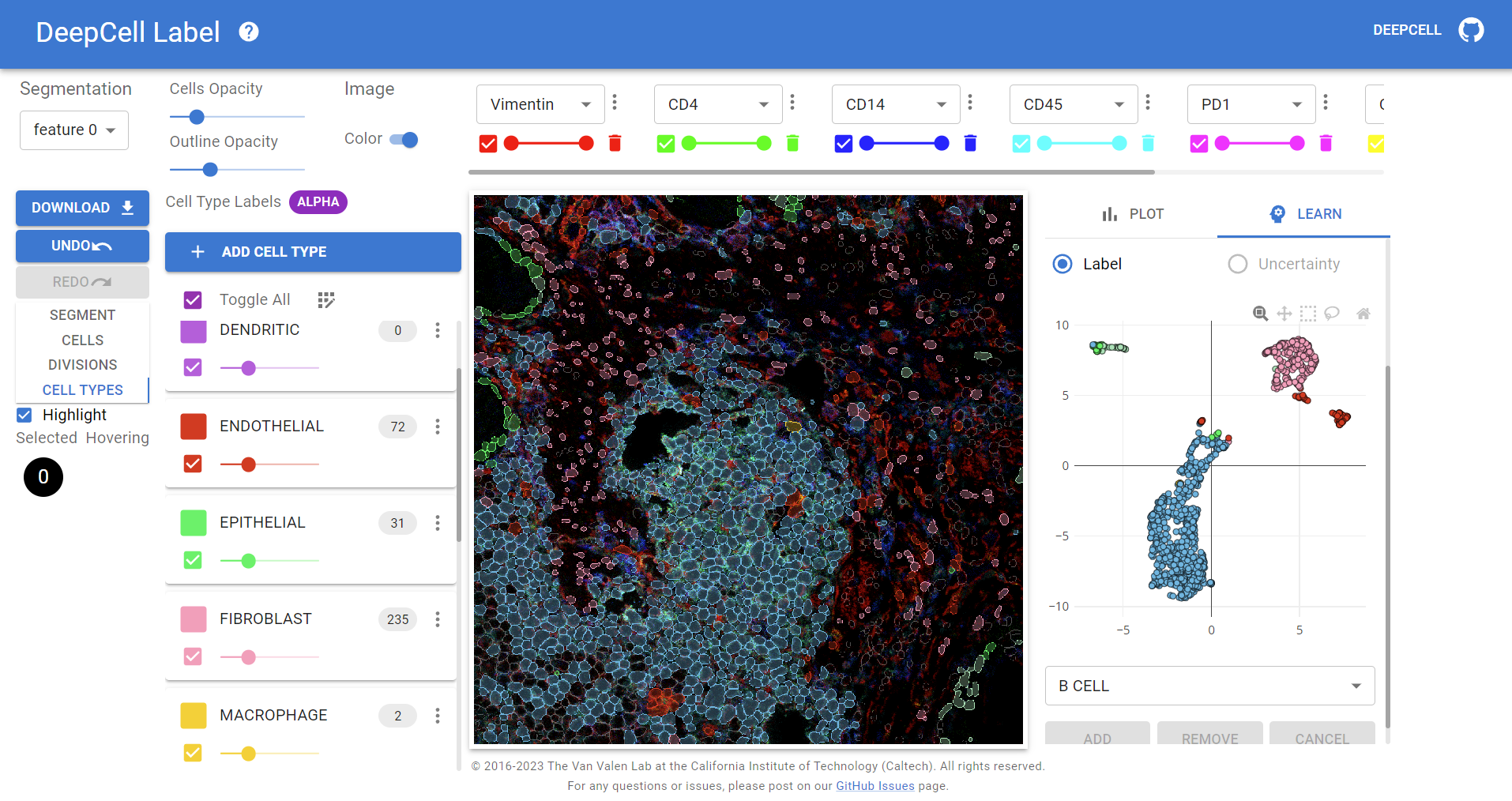
How can we obtain large amounts of rich biological image data for deep learning pipelines? Tailor-made software for data annotation is crucial for complex human-in-the-loop frameworks. DeepCell Label is a cloud-based web application built on Flask and React that allows anyone to remotely annotate biological image data with segmentation masks, division events, spots, and cell types. As a Software Engineer in the Van Valen Lab, I have been the primary developer and maintainer of DeepCell Label for the past year and added functionality for interactive cell type annotation, on-the-fly marker expression analyses, and in-browser training using Tensorflow.js for active learning accelerated labeling.
CipherDB
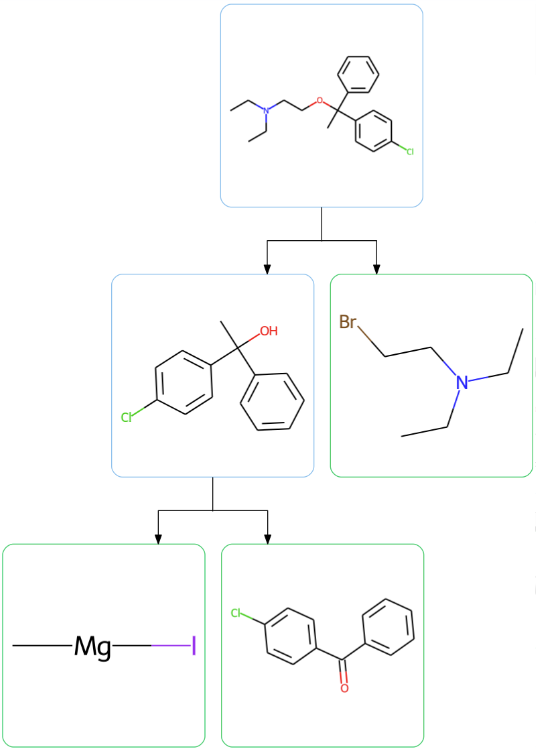
Computational tools such as generative models have shown promise in proposing molecules with highly desirable properties (eg. for drug discovery). The 2020 NCATS ASPIRE Challenge sought approaches to tackling the opioid crisis through discovery of analgesics with non-addictive properties. As part of my NSF REU Summer Research internship with Prof. Gaurav Chopra at Purdue, I wrote MongoDB triggers to automatically calculate synthesis-related metrics and visualize proposed retrosynthetic routes to any drug-like molecule that was added to a database, including those output by generative models. I also developed a new synthetic feasibility metric by using a feed-forward neural network to predict the number of steps required to synthesize a given molecule as proposed by a CASP tool (ASKCOS). Our work on the database and associated platforms went on to win the grand prize in the challenge.
